Reading
frames and ORFs
Objectives of this
assgnment:
Describe what the terms reading frames and ORFs mean.
Describe how how potential ORFs are detected.
Describe what is meant by template and sense strands of DNA.
Describe fundamental differences between prokaryotic-like
genomes (mitochondrial in this case) and typical eukarytotic
chromosomal genes;
Understand that the "universal" genetc code is not 100%
universal.
Be able to use ORF Finder and BLAST programs and to interpret findings from them.
Cellular DNA is a
double-stranded,anti-parallel
molecule that contains genes that are both transcribed and
translated (protein-coding genes), genes that are only
transcribed to RNA (e.g. tRNA, rRNA, snRNA),
and
non-coding regions not generally transcribed (e.g., regulatory
sequences, centromeres).
Either of the two
strands can contain genes. When transcribed, the template (noncoding,
antisense, minus) strand
of DNA is complementary to the RNA, and the sense (coding, plus) strand of the
DNA has a sequence identical to RNA in orientation (5' to 3')
and in sequence (if you replace the uridines
of the RNA with thymidines in the
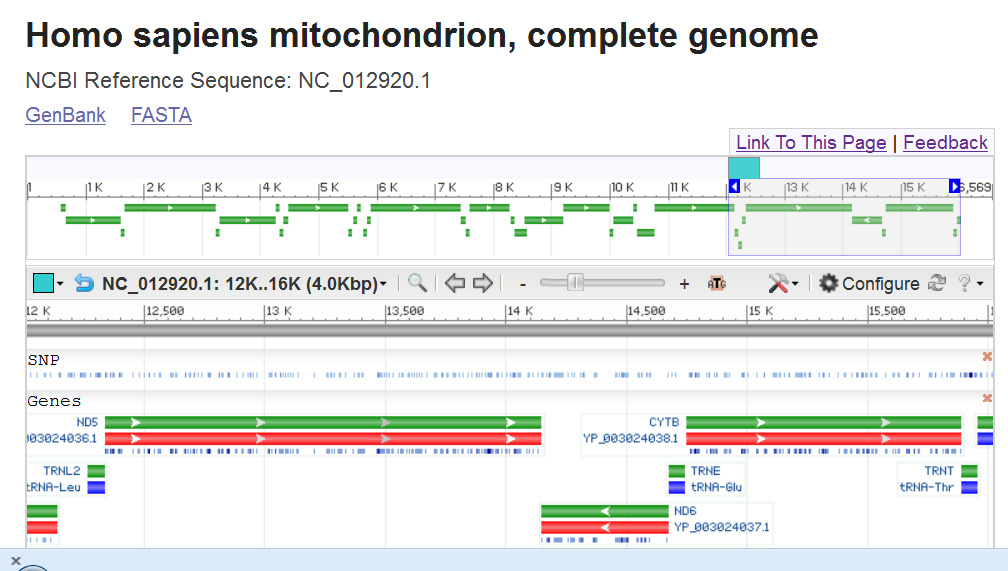
DNA). Below is a figure from part of the human mitochondrial
genome (NCBI- 2008 image of mapview; current version a bit
different in format). This image shows a region
containing two genes, ND5 and ND6,
that encode proteins that are part of the electron transport
chain, and for four different tRNAs.
The little arrows next to the gene symbol indicate the
orientation of the sense strand.
Q1. Compare ND5 and ND6 genes on the
above map regarding coding strands and size of the
gene.
Genes that code for proteins are also known as open reading frames,
or ORFs. ORFs begin with a start codon (usually
ATG in the sense strand of the DNA sequence)and
end with one of three stop codons.
Searching for ORFs in a DNA
sequence requires analysis of both strands, searching for
start codons from each 5' end.
Once a putative start codon is found, the sequence is
read as triplet codons until a
stop codon is reached. Each strand can be read in this way in
3 possible triplet reading
frames, as illustrated below. Since the DNA is
double-stranded, 6 reading frames need to be scanned for ORFs, 3 for each strand.
For the
sequence: 5' cgggtgatctcata 3'
reading frame 1 cgg gtg
atc tca
ta
reading frame 2 c ggg tga
tct cat a
reading frame 3 cg ggt gat ctc
ata
And for the complementary
sequence: 5' tatgagatcacccg 3'
reading frame 1 tat gag atc acc cg
reading frame 2 t atg
aga tca
ccc g
Q2. Which of the above examples
has a potential start of an ORF?
As you might expect,
searching for ORFs in long
stretches of DNA sequences is pretty tedious. Fortunately,
there exist tools for this type of analysis. The NCBI Home Page at http://www.ncbi.nlm.nih.gov
has numerous tools for analyzing DNA sequences, including a
program called ORF finder. The link for the program is
toward the bottom of the NCBI main page under
resources/sequence analysis, in the second portion of the
list, Tools. Alternatively, from the NCBI home page click on
Resources A-Z on the left and find ORF finder in the
alphabetical listing. Since they have
hidden things in their new page format, you may find it
useful to bookmark some of these commonly used tools in your
web browser. To
run the program copy and paste the sequence of interest into
the large data input window that's labeled "sequence in FASTA format". Click on the "OrfFind" button above the
window. Within a minute an ORF list will be
shown. On the right will be the ORFs
listed by length. On the left will be a map of the DNA
sequence showing the ORFs (in
blue) in each of the 6 possible reading frames.
In this exercise you will examine two different gene
sequences with ORF finder.
Gene located on
human mitochondrial DNA
Select and copy the sequence below (Ctrl +c) and
paste (Ctrl +v) into ORF finder. Since this is a
mitochondrial gene, the default codons
will not work! Select vertebrate mitochondrial from the choices at the bottom of the
ORF finder. Click on the "OrfFind"
button above the window. Within a minute an ORF list
will be shown. On the left will be a map
of the DNA sequence showing the ORFs
(in blue) in each of the 6 possible reading frames. On the
right will be the ORFs listed
by length. Only one ORF should be present. Click
on it examine the sequence.
Q3. Which
reading frame contains the ORF (+1, -2 etc.) How long is the
protein? At which base pair (number in the DNA
sequence) does the ORF start? At which base pair
(number in the DNA sequence) does the ORF end? What
start codon is used? What stop codon is used? What are
the first and last amino acids in the sequence? Any
surprises?
Q4. If we had included in our DNA sequence the
sequence for ND6, which of the reading frames (+1,+2, +3) or
(-1, -2, -3) would you expect the ND6 ORF to be in?
Another tool you can access through ORF Finder is a
direct link to BLAST searches. BLAST (Basic Local
Alignment Search Tool) is a program which searches
sequences at GenBank for
matches to an input sequence. The link is directly
above the maps showing the ORFs.
There are several varieties of BLAST searches,
including searches that match nucleotide sequences, protein
sequences, and searches that will match a protein sequence
to DNA sequences "translated" by the BLAST program.
Since we are working with a translated DNA sequence, we will
use the indicated default BLAST (blastp)
from the site which matches your input protein sequence (the
translated ORF) with the "translated" nucleotide database
(nr).
To run the program, make
sure the ORF you want to analyze is
highlighted, then simply click on the BLAST
button. This will open a new page that has a request
ID and additional information regarding format. Next
to the view report button check the box for “Show results in
a new window”, and then click on view report. It may
take a minute or so for the full report to show up. At the top of the report
you will see a map showing conserved domains in the protein
detected by an additional matching program. Below that you will
see a table summarizing alignment “hits”. Below the table is
a list of matching sequences. The higher the score,
the better the match; the smaller the E value, the less
likely the match is just by chance. The E values are
comparable to the “p value” you would get in a statistical
test, except that they can be >1.
Q4.
Look at the first match. What does it correspond to?
What is the E value for the match?
Open the top link
to AAK17750.1 to examine the
GenBank entry. In the top
portion of the entry you will find a summary of information.
Q5. What is the name (definition) of the entry? How was the
protein sequence obtained?
>mitochondrial sequence GI|13273141:12340..14151
TAATAACCATGCACACTACTATAACCACCCTAACCCTGACTTCCCTAATTCCCCCCATCCTTACCACCCTCG
TTAACCCTAACAAAAAAAACTCATACCCCCATTATGTAAAATCCATTGTCGCATCCACCTTTATTATCAG
TCTCTTCCCCACAACAATATTCATGTGCCTAGACCAAGAAGTTATTATCTCGAACTGACACTGAGCCACA
ACCCAAACAACCCAGCTCTCCCTAAGCTTCAAACTAGACTACTTCTCCATAATATTCATCCCTGTAGCAT
TGTTCGTTACATGGTCCATCATAGAATTCTCACTGTGATATATAAACTCAGACCCAAACATTAATCAGTT
CTTCAAATATCTACTCATTTTCCTAATTACCATACTAATCTTAGTTACCGCTAACAACCTATTCCAACTG
TTCATCGGCTGAGAGGGCGTAGGAATTATATCCTTCTTGCTCATCAGTTGATGATACGCCCGAGCAGATG
CCAACACAGCAGCCATTCAAGCAGTCCTATACAACCGTATCGGCGATATCGGTTTCATCCTCGCCTTAGC
ATGATTTATCCTACACTCCAACTCATGAGACCCACAACAAATAGCCCTTCTAAACGCTAATCCAAGCCTC
ACCCCACTACTAGGCCTCCTCCTAGCAGCAGCAGGCAAATCAGCCCAATTAGGTCTCCACCCCTGACTCC
CCTCAGCCATAGAAGGCCCCACCCCAGTCTCAGCCCTACTCCACTCAAGCACTATAGTTGTAGCAGGAAT
CTTCTTACTCATCCGCTTCCACCCCCTAGCAGAAAATAGCCCACTAATCCAAACTCTAACACTATGCTTA
GGCGCTATCACCACTCTGTTCGCAGCAGTCTGCGCCCTTACACAAAATGACATCAAAAAAATCGTAGCCT
TCTCCACTTCAAGTCAACTAGGACTCATAATAGTTACAATCGGCATCAACCAACCACACCTAGCATTCCT
GCACATCTGTACCCACGCCTTCTTCAAAGCCATACTATTTATGTGCTCCGGGTCCATCATCCACAACCTT
AACAATGAACAAGATATTCGAAAAATAGGAGGACTACTCAAAACCATACCTCTCACTTCAACCTCCCTCA
CCATTGGCAGCCTAGCATTAGCAGGAATACCTTTCCTCACAGGTTTCTACTCCAAAGACCACATCATCGA
AACCGCAAACATATCATACACAAACGCCTGAGCCCTATCTATTACTCTCATCGCTACCTCCCTGACAAGC
GCCTATAGCACTCGAATAATTCTTCTCACCCTAACAGGTCAACCTCGCTTCCCCACCCTTACTAACATTA
ACGAAAATAACCCCACCCTACTAAACCCCATTAAACGCCTGGCAGCCGGAAGCCTATTCGCAGGATTTCT
CATTACTAACAACATTTCCCCCGCATCCCCCTTCCAAACAACAATCCCCCTCTACCTAAAACTCACAGCC
CTCGCTGTCACTTTCCTAGGACTTCTAACAGCCCTAGACCTCAACTACCTAACCAACAAACTTAAAATAA
AATCCCCACTATGCACATTTTATTTCTCCAACATACTCGGATTCTACCCTAGCATCACACACCGCACAAT
CCCCTATCTAGGCCTTCTTACGAGCCAAAACCTGCCCCTACTCCTCCTAGACCTAACCTGACTAGAAAAG
CTATTACCTAAAACAATTTCACAGCACCAAATCTCCACCTCCATCATCACCTCAACCCAAAAAGGCATAA
TTAAACTTTACTTCCTCTCTTTCTTCTTCCCACTCATCCTAACCCTACTCCTAATCACATAATAA
Gene located on
human chromosome 11
When you run ORF
finder on this sequence, you will find several small ORFs, only a some of which are real
protein coding regions for the gene. The N terminus of
the protein is found in reading frame +3: 51-236.
Subsequent relevant portions of the protein are in reading
frames +1:115-516, and +1:1261-1449.
The C terminus is in reading frame +2, 1361-1474.
Select these ORFs and
use the BLAST program to the find the matches. Examine
the alignments. Open up and read one of the GenBank descriptions of the
protein.
Q7. Note the overlap of the calculated ORF sequences +3:51-236 and +1:115-516. Is this something you normally expect in an expressed gene? Examine the BLAST alignments for these regions. Do the calculated ORF protein sequences (query) entirely correspond to the beta globin protein sequence (subject)?
Q8. Different
portions of the beta globin protein sequence are
found in the +1, +2 and +3 reading frames?
Why wouldn't you expect beta globin ORFs to be in the -1,-2,-3
reading frames?
Q9. Put it all together--What
fundamental difference in gene
structure are we observing when we compare the
discontinuous beta globin gene on chromosome 11 with
the simple ORF pattern from the mitochondrial gene?
Q10. How would the ORF pattern differ
if we could run ORF finder on the mRNA sequence of this
gene?
>gi|224589802:c5248301-5246696 Homo sapiens chromosome 11 ACATTTGCTTCTGACACAACTGTGTTCACTAGCAACCTCAAACAGACACCATGGTGCATCTGACTCCTGA GGAGAAGTCTGCCGTTACTGCCCTGTGGGGCAAGGTGAACGTGGATGAAGTTGGTGGTGAGGCCCTGGGC AGGTTGGTATCAAGGTTACAAGACAGGTTTAAGGAGACCAATAGAAACTGGGCATGTGGAGACAGAGAAG ACTCTTGGGTTTCTGATAGGCACTGACTCTCTCTGCCTATTGGTCTATTTTCCCACCCTTAGGCTGCTGG TGGTCTACCCTTGGACCCAGAGGTTCTTTGAGTCCTTTGGGGATCTGTCCACTCCTGATGCTGTTATGGG CAACCCTAAGGTGAAGGCTCATGGCAAGAAAGTGCTCGGTGCCTTTAGTGATGGCCTGGCTCACCTGGAC AACCTCAAGGGCACCTTTGCCACACTGAGTGAGCTGCACTGTGACAAGCTGCACGTGGATCCTGAGAACT TCAGGGTGAGTCTATGGGACGCTTGATGTTTTCTTTCCCCTTCTTTTCTATGGTTAAGTTCATGTCATAG GAAGGGGATAAGTAACAGGGTACAGTTTAGAATGGGAAACAGACGAATGATTGCATCAGTGTGGAAGTCT CAGGATCGTTTTAGTTTCTTTTATTTGCTGTTCATAACAATTGTTTTCTTTTGTTTAATTCTTGCTTTCT TTTTTTTTCTTCTCCGCAATTTTTACTATTATACTTAATGCCTTAACATTGTGTATAACAAAAGGAAATA TCTCTGAGATACATTAAGTAACTTAAAAAAAAACTTTACACAGTCTGCCTAGTACATTACTATTTGGAAT ATATGTGTGCTTATTTGCATATTCATAATCTCCCTACTTTATTTTCTTTTATTTTTAATTGATACATAAT CATTATACATATTTATGGGTTAAAGTGTAATGTTTTAATATGTGTACACATATTGACCAAATCAGGGTAA TTTTGCATTTGTAATTTTAAAAAATGCTTTCTTCTTTTAATATACTTTTTTGTTTATCTTATTTCTAATA CTTTCCCTAATCTCTTTCTTTCAGGGCAATAATGATACAATGTATCATGCCTCTTTGCACCATTCTAAAG AATAACAGTGATAATTTCTGGGTTAAGGCAATAGCAATATCTCTGCATATAAATATTTCTGCATATAAAT TGTAACTGATGTAAGAGGTTTCATATTGCTAATAGCAGCTACAATCCAGCTACCATTCTGCTTTTATTTT ATGGTTGGGATAAGGCTGGATTATTCTGAGTCCAAGCTAGGCCCTTTTGCTAATCATGTTCATACCTCTT ATCTTCCTCCCACAGCTCCTGGGCAACGTGCTGGTCTGTGTGCTGGCCCATCACTTTGGCAAAGAATTCA CCCCACCAGTGCAGGCTGCCTATCAGAAAGTGGTGGCTGGTGTGGCTAATGCCCTGGCCCACAAGTATCA CTAAGCTCGCTTTCTTGCTGTCCAATTTCTATTAAAGGTTCCTTTGTTCCCTAAGTCCAACTACTAAACT GGGGGATATTATGAAGGGCCTTGAGCATCTGGATTCTGCCTAATAAAAAACATTTATTTTCATTGC