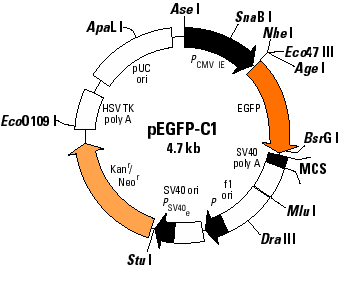
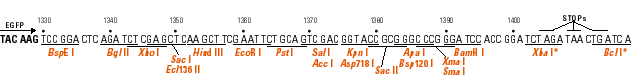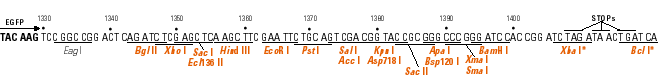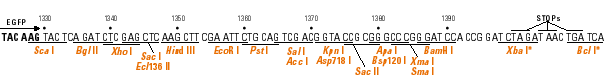The goal of this exercise is to become familiar with a technique commonly used in recombinant DNA technology - the subcloning of a cloned DNA into a specialized vector. You will use several Web tools to aid you in this project.
In this specific case you want to create a fusion protein between your protein and green fluorescent protein. The vector you will be using comes in 3 forms, each with a different reading frame in the multiple cloning site. One of these vectors will allow in frame fusion of the GFP at the N terminus of your protein. Your mission is to determine which vector and which restriction enzyme(s) to use.
Links you will find useful:
ORF finder: a tool at NCBI that will detect open reading frames in a piece of DNA
BLAST: Basic local alignment of sequences tool, maintained by NCBI. This tool will search GenBank sequences for a match to your query, and generate alignments with the matches. There are a number of forms of BLAST. You will want to use Nucleotide-nucleotide BLAST (blastn) for this search.
NEB Cutter: a tool provided by New England Biolabs, a company best known for its restriction enzymes, that will analyze a DNA sequence and detect restriction sites. By clicking on the line representing the DNA sequence you can select areas to zoom in upon in order to see the actual sequence. It will also find ORFs for you, and will also link you to protein BLAST for identification of the sequence.
Steps to take:
Identify the cloned sequence using BLAST
Determine where the ORF is in relation to the bases. The alignment of codons and amino acids from ORF finder will be useful.
Determine the restriction sites available for cleaving out the DNA from its current vector. You can choose two unque enzymes flanking the insert, if available, or choose a single enzyme that excises the insert. These restriction sites need to be available in the same orientation, 5' to 3' of the sense strand, in the GFP fusion vector; i.e., don't put the DNA in backwards!
Now determine which of the three vectors has the same reading frame as the insert. The easiest approach is to look at the codons in relation to the restriction sites.
gccaagcttg catgcctgca ggtcgactct agaggaaccc cgggtaccga gctcgaattc cgtactccgc catgaccacc gctgtcaccg ccgctgtttc tttcccctct accaaaacca
gggaagacaa gaatggaaag ccccataagt tgagattgta ctcgatcgcc agcagtgctc
atgacgctgg agagacgatc aagggagtct gctccaactt cttgtgtgac ttgaaacccg
acgcgacaat tatcatgctt ggaactggaa cggggattgc tcctttccgt tcattcttgt
tgggtgtacc cacaagcagt tctcttctct acaaagagga atttgagaag atgaaggaaa
gggagaagat gtacattcaa acccgaatgg cacaatacgc agttgagcta tgggaaatgt
acgacattat ggtttcattg gctgctgcag aaggcattga ttggattgaa tacaagaggc
ccttattcct ttttgtgaag catcttctat atttatctat tgccattatt atctcactgc